Part:BBa_K3762010
YFP_LOV driven by T7 promoter
A generator using the well chararcterized "superfolder GFP driven by T7 promoter" BBa_I746909 where the sfGFP is replaced with the YFP_LOV from GO_Paris-Saclay BBa_K3427000
We wanted to modify an existing part where the improvement was in its added utility so that it would fit the needs of future teams whose project has aspects that require it to work under hypoxic conditions or where the availability of molecular oxygen is low.
Our chosen part was the well characterized signal generator “superfolder GFP driven by T7 promoter” BBa_I746909 by Cambridge 2007 in which we replaced its sfGFP with the “heat stable fluorescent YFP_LOV protein” BBa_K3427000 from GO_Paris-Saclay’s 2020 team.
Characterization
The linear fragment of BBa_K3762010 was cloned using Gibson assembly into a pENZ004 plasmid provided by the PhotoSyn lab at NTNU. We proceeded to transform the plasmid into our E. coli BL21 cells and selected using ampicillin resistance.
Fluorescence measurement settings using a Tecan Infinite 200 Pro:
| Setting | Value |
| Excitation wavelength | 470 nm |
| Emission wavelength | 520 nm |
| Excitation Bandwidth | 9 nm |
| Emission Bandwidth | 20 |
| Gain | 100 |
| Number of flashes | 22 |
| Integration Time | 20 us |
| Lag time | 0 us |
| Settle time | 0 ms |
| Z-position | 20000 um |
| Setting | Value |
| Excitation Wavelength | 488 nm |
| Emission Wavelength | 536 nm |
| Excitation Bandwidth | 9 nm |
| Emission Bandwidth | 20 |
| Gain | 100 |
| Number of flashes | 22 |
| Integration Time | 20 us |
| Lag time | 0 us |
| Settle time | 0 ms |
| Z-position | 20000 um |
Results
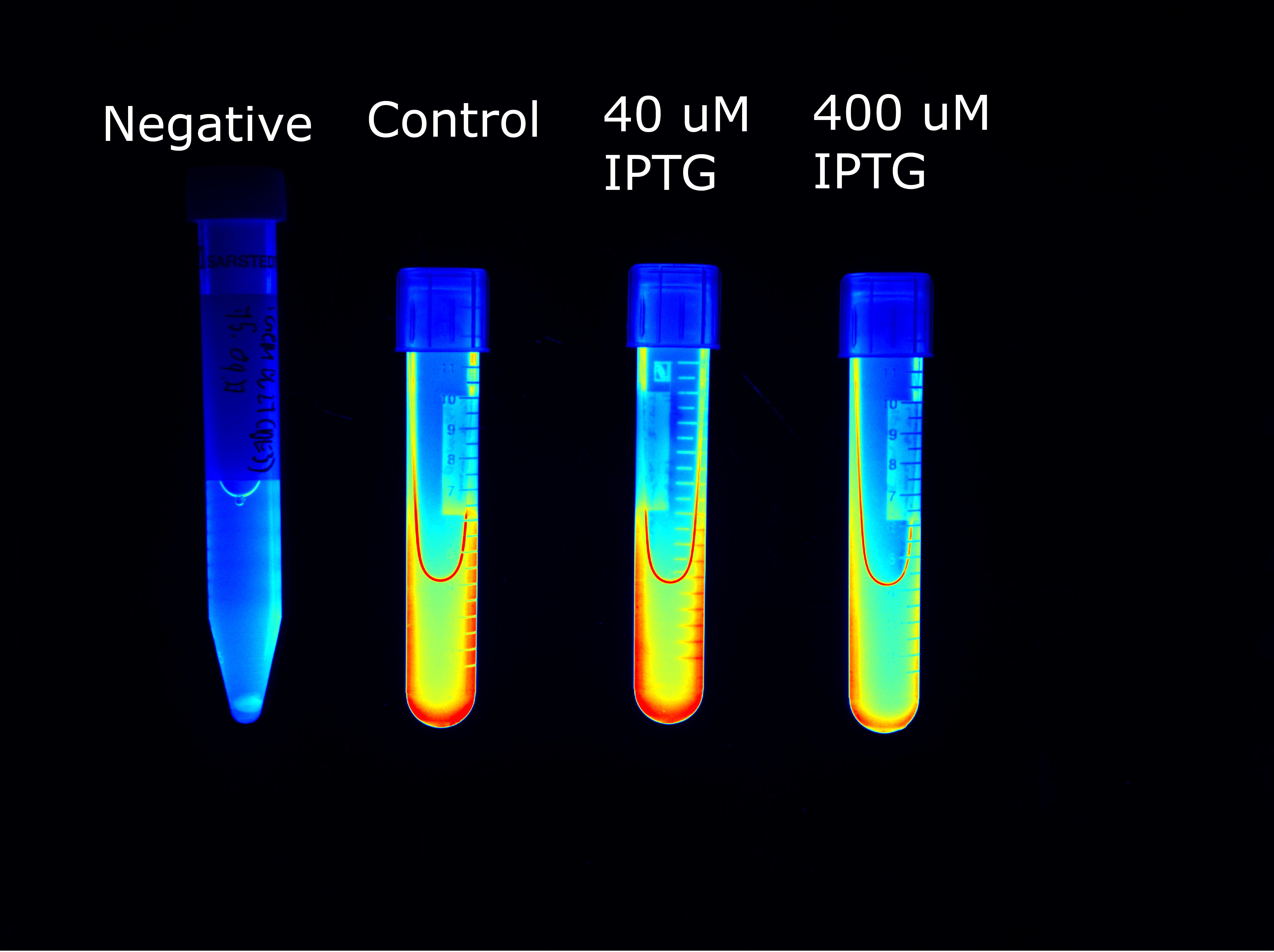
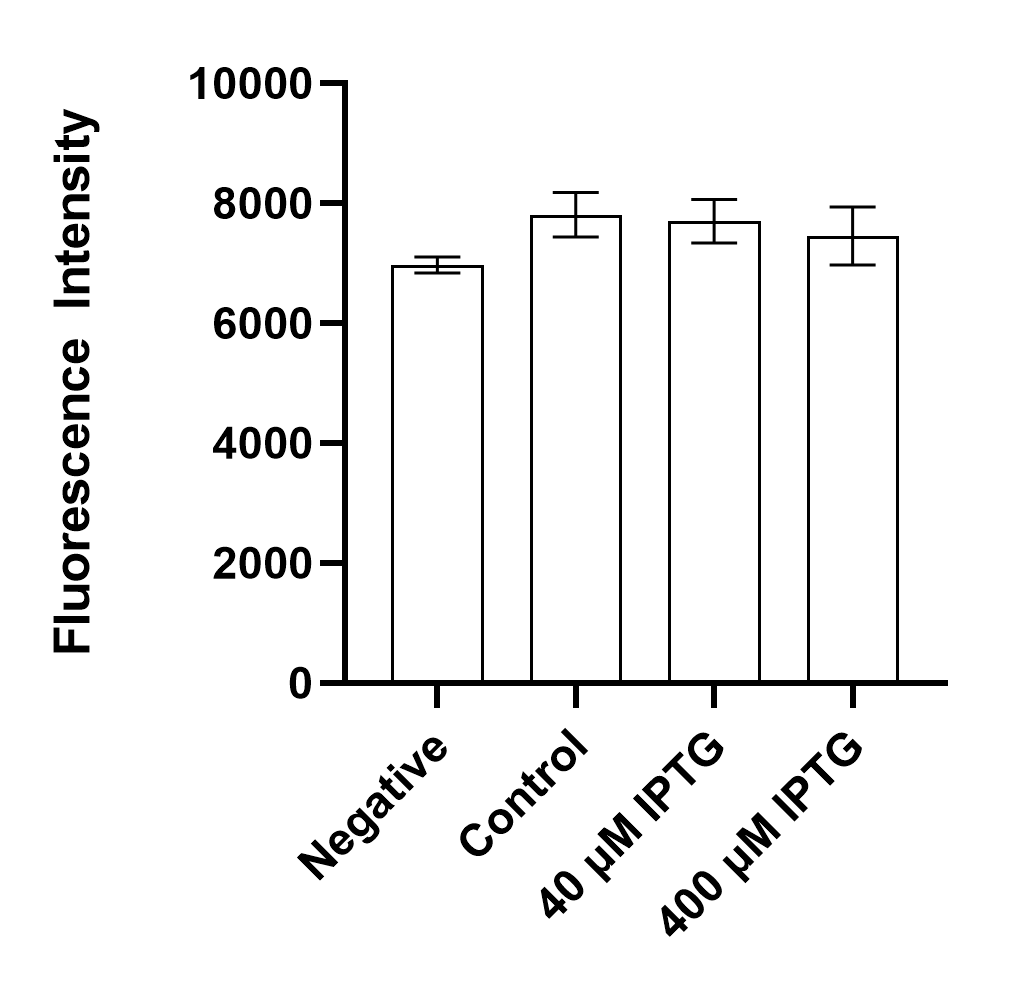
| Sample | Plasmid | Inducer (IPTG) | Fluorescence |
| Negative | No | No | No |
| Control | Yes | No | Yes |
| 40 µM IPTG | Yes | Yes | Yes |
| 400 µM IPTG | Yes | Yes | Yes |

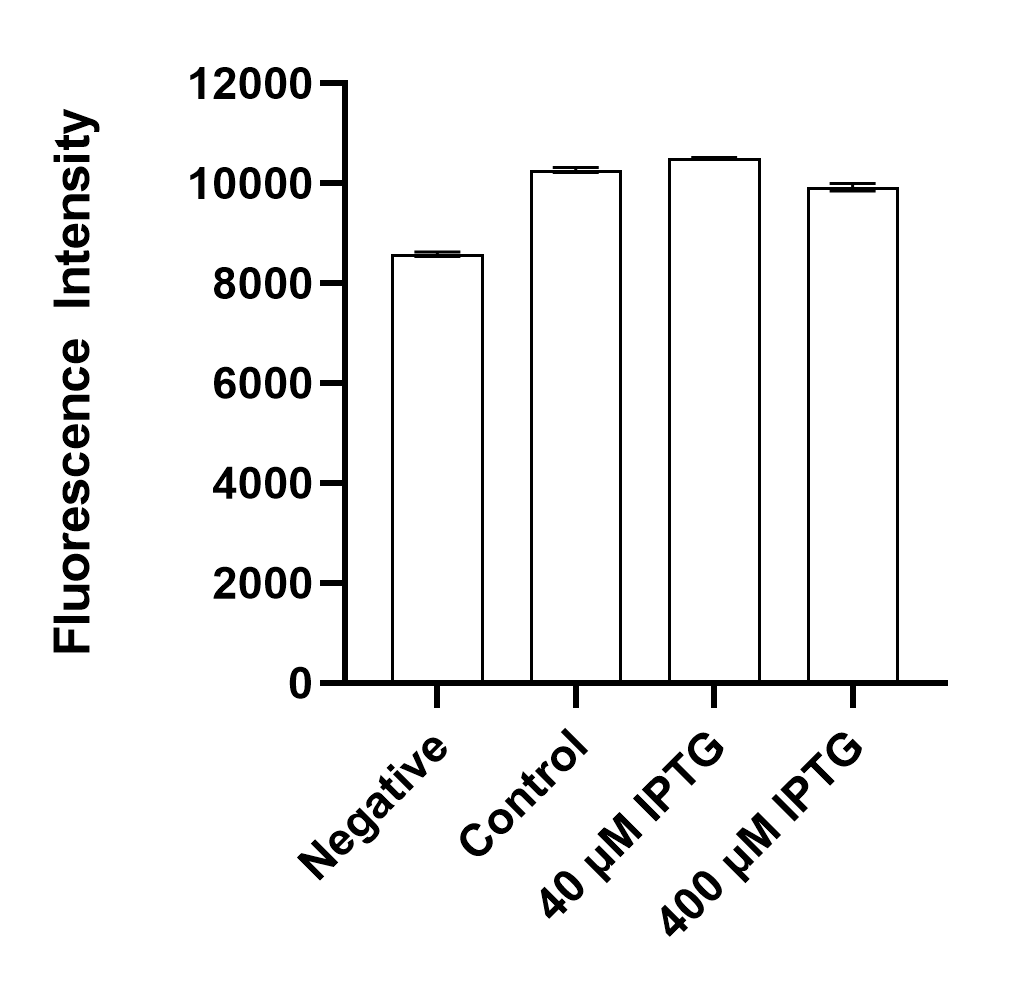
| Sample | Plasmid | Inducer (IPTG) | Fluorescence |
| Negative | No | No | No |
| Control | Yes | No | Yes |
| 40 µ IPTG | Yes | Yes | Yes |
| 400 µ IPTG | Yes | Yes | Yes |
Sequencing results from Eurofins confirmed successful insert of plasmids with each respective construct.
Conclusion
Sequencing confirmed successful insert in the pENZ0004 plasmid and results indicate that we were able to clone our construct into our E. coli BL21 cells.
Results indicate that BBa_K3762010 is not as fluorescent as BBa_I746909 but does exhibit similar behavior with and without inducers in our inducer concentration range [40 - 400 µ IPTG].
Due to how flavin-based fluorescent protein does not require molecular oxygen to attain its fluorescent form, the case for the improvement over sfGFP lies in its function in hypoxic conditions. Potential applications can be in microfluidics or a diagnostic setting where the environment has low or none molecular oxygen availability.

Figure 7: Plasmid map of the pENZ004 plasmid and our construct BBa_k3762010.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References:
[1] Limitations of the Reporter Green Fluorescent Protein under Simulated Tumor Conditions
Claudia Coralli, Maja Cemazar, Chryso Kanthou, Gillian M. Tozer and Gabi U. Dachs
Cancer Res June 15 2001 (61) (12) 4784-4790;
https://cancerres.aacrjournals.org/content/61/12/4784
[2] The photophysics of LOV-based fluorescent proteins - new tools for cell biology
Marcus Wingen, Janko Potzkei Stephan Endres, Giorgia Casini, Christian Rupprecht, Christoph Fahlke Ulrich Krauss, Karl-Erich Jaeger, Thomas Drepper and Thomas Gensch
Photochem. Photobiol. Sci., 2014,13, 875-883
https://doi.org/10.1039/C3PP50414J
[3] A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: a framework for ultra-high resolution structural studies Nazarenko et al. 2019, Photochem
Photochem. Photobiol. Sci., 2019,18, 1793-1805
| None |
